xhca
Version 1.0
Fonctions :
xhca is a program used to create H.C.A. (Hydrophobic
Cluster Analysis) diagrams from the sequence of a protein.
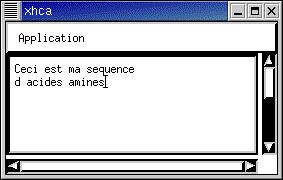 |
The user enter the sequence
in a window by typing it from the
keyboard or using copy/paste |
Then he chooses one of the available plot type :
-
standard plot : (recommanded value)
In this case only black/white or color options and the title
are available.
This type of plot is very similar to the standard one;
only its "look" is slightly different.
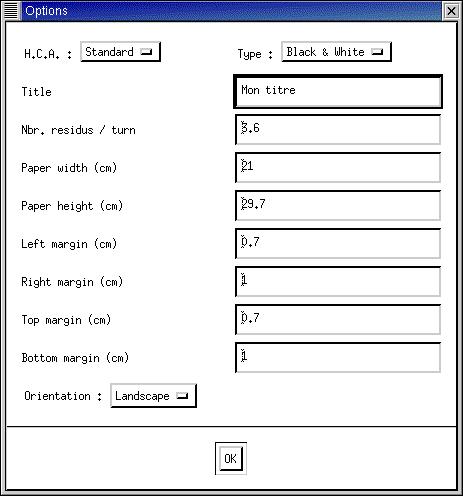 |
In this case options for the
orientation, the size of the paper
and the margins are available.
|
enable to choose the number of residus per turn,
for helices that are not alpha.
In this case "mosaics" are not drawn and Proline
does not break hydrophobic clusters.
Finally the user chooses a filename to save
the sequence:
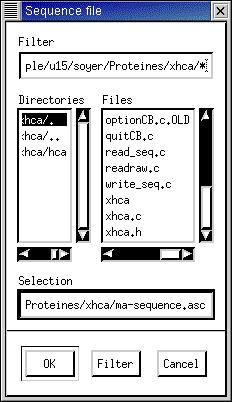 |
for example
ma-sequence.asc (ou .seq)
and the plot will be done in a
PostScript file named
ma-sequence.ps |
Mandatory ressources :
Unix workstation (or Linux PC, or Mac under MacOS 10) with a
C compiler, and the X-Window and Motif libraries.
X-Window screen or X terminal.
Distribution :
Sources of the program are freely available (without guaranty)
by ftp, in the form of a compressed tar file :
xhca1.0.tar.Z
from the external network, or
xhca1.0.tar.Z
from the local network.
Installation :
Uncompress the tar file : "uncompress xhca1.0.tar.Z"
extract the files : "tar -xvf xhca1.0.tar"
in the directory xhca/hca modify the makefile to adapt it
to your machine, then execut it :
"cd xhca/hca"
"vi makefile" ...
"make"
in the dircetory xhca modify the makefile to adapt it
to your machine, then execut it:
"cd .."
"vi makefile" ...
"make"



